Working with the EXIOBASE EE MRIO database¶
Getting EXIOBASE¶
EXIOBASE 1 (developed in the fp6 project EXIOPOL) and EXIOBASE 2 (outcome of the fp7 project CREEA) are available on http://www.exiobase.eu
You need to register before you can download the full dataset.
### EXIOBASE 1
To download EXIOBASE 1 for the use with pymrio, navigate to https://www.exiobase.eu - tab “Data Download” - “EXIOBASE 1 - full dataset” and download either
- pxp_ita_44_regions_coeff_txt for the product by product (pxp) MRIO system or
- ixi_fpa_44_regions_coeff_txt for the industry by industry (ixi) MRIO system or
- pxp_ita_44_regions_coeff_src_txt for the product by product (pxp) MRIO system with emission data per source or
- ixi_fpa_44_regions_coeff_src_txt for the industry by industry (ixi) wMRIO system with emission data per source.
The links above directly lead to the required file(s), but remember that you need to be logged in to access them.
The pymrio parser works with the compressed (zip) files as well as the unpacked files. If you want to unpack the files, make sure that you store them in different folders since they unpack in the current directory.
EXIOBASE 2¶
EXIOBASE 2 is available at ttp://www.exiobase.eu - tab “Data Download” - “EXIOBASE 2 - full dataset”. You can download either
- MrIOT PxP ita coefficient version2 2 2 for the product by product (pxp) MRIO system or
- MrIOT IxI fpa coefficient version2 2 2 for the industry by industry (ixi) MRIO system.
The links above directly lead to the required file(s), but remember that you need to be logged in to access them.
The pymrio parser works with the compressed (zip) files as well as the unpacked files. You can unpack the files together in one directory (unpacking creates a separate folder for each EXIOBASE 2 version). The unpacking of the PxP version also creates a folder “__MACOSX” - you can delete this folder.
EXIOBASE 3¶
EXIOBASE 3 is currently not publicly available. However, pymrio already includes a parser for the preliminary version. If you have access to this version, you can download the files as provided and use the preliminary pymrio exiobase3 parser. Manually adjustment might be needed depending on the available sub-version of EXIOBASE 3.
Parsing¶
In [1]:
import pymrio
For each publically available version of EXIOBASE pymrio provides a specific parser. To parse EXIOBASE 1 use:
In [2]:
exio1 = pymrio.parse_exiobase1(path='/tmp/mrios/exio1/121016_EXIOBASE_pxp_ita_44_regions_coeff_txt.zip')
The parameter ‘path’ needs to point to either folder with the extracted EXIOBASE1 files for the downloaded zip file.
Similarly, EXIOBASE2 can be parsed by:
In [3]:
exio2 = pymrio.parse_exiobase2(path='/tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip',
charact=True, popvector='exio2')
The additional parameter ‘charact’ specifies if the characterization matrix provided with EXIOBASE 2 should be used. This can be specified with True or False; in addition, a custom one can be provided. In the latter case, pass the full path to the custom characterisatio file to ‘charact’.
The parameter ‘popvector’ allows to pass information about the population per EXIOBASE2 country. This can either be a custom vector of, if ‘exio2’ is passed, the one provided with pymrio.
For the rest of the tutorial, we use exio2; deleting exio1 to free some memory:
In [4]:
del exio1
Exploring EXIOBASE¶
After parsing a EXIOBASE version, the handling of the database is the same as for any IO. Here we use the parsed EXIOBASE2 to explore some characteristics of the EXIBOASE system.
After reading the raw files, metadata about EXIOBASE can be accessed within the meta field:
In [5]:
exio2.meta
Out[5]:
Description: Metadata for pymrio
MRIO Name: EXIOBASE
System: pxp
Version: 2.2.2
File: None
History:
20180111 10:25:01 - FILEIO - EXIOBASE data FY_materials parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFDMaterials_version2.2.2.txt
20180111 10:25:01 - FILEIO - EXIOBASE data FY_emissions parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFDEmissions_version2.2.2.txt
20180111 10:25:01 - FILEIO - EXIOBASE data S_resources parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrResources_version2.2.2.txt
20180111 10:25:00 - FILEIO - EXIOBASE data S_materials parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrMaterials_version2.2.2.txt
20180111 10:24:59 - FILEIO - EXIOBASE data S_emissions parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrEmissions_version2.2.2.txt
20180111 10:24:58 - FILEIO - EXIOBASE data S_factor_inputs parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFactorInputs_version2.2.2.txt
20180111 10:24:58 - FILEIO - EXIOBASE data Y parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFinalDemand_version2.2.2.txt
20180111 10:24:57 - FILEIO - EXIOBASE data A parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrIot_version2.2.2.txt
Custom points can be added to the history in the meta record. For example:
In [6]:
exio2.meta.note("First test run of EXIOBASE 2")
exio2.meta
Out[6]:
Description: Metadata for pymrio
MRIO Name: EXIOBASE
System: pxp
Version: 2.2.2
File: None
History:
20180111 10:25:02 - NOTE - First test run of EXIOBASE 2
20180111 10:25:01 - FILEIO - EXIOBASE data FY_materials parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFDMaterials_version2.2.2.txt
20180111 10:25:01 - FILEIO - EXIOBASE data FY_emissions parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFDEmissions_version2.2.2.txt
20180111 10:25:01 - FILEIO - EXIOBASE data S_resources parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrResources_version2.2.2.txt
20180111 10:25:00 - FILEIO - EXIOBASE data S_materials parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrMaterials_version2.2.2.txt
20180111 10:24:59 - FILEIO - EXIOBASE data S_emissions parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrEmissions_version2.2.2.txt
20180111 10:24:58 - FILEIO - EXIOBASE data S_factor_inputs parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFactorInputs_version2.2.2.txt
20180111 10:24:58 - FILEIO - EXIOBASE data Y parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrFinalDemand_version2.2.2.txt
20180111 10:24:57 - FILEIO - EXIOBASE data A parsed from /tmp/mrios/exio2/mrIOT_PxP_ita_coefficient_version2.2.2.zip/mrIOT_PxP_ita_coefficient_version2.2.2/mrIot_version2.2.2.txt
To check for sectors, regions and extensions:
In [7]:
exio2.get_sectors()
Out[7]:
Index(['Paddy rice', 'Wheat', 'Cereal grains nec', 'Vegetables, fruit, nuts',
'Oil seeds', 'Sugar cane, sugar beet', 'Plant-based fibers',
'Crops nec', 'Cattle', 'Pigs',
...
'Paper for treatment: landfill',
'Plastic waste for treatment: landfill',
'Inert/metal/hazardous waste for treatment: landfill',
'Textiles waste for treatment: landfill',
'Wood waste for treatment: landfill',
'Membership organisation services n.e.c.',
'Recreational, cultural and sporting services', 'Other services',
'Private households with employed persons',
'Extra-territorial organizations and bodies'],
dtype='object', name='sector', length=200)
In [8]:
exio2.get_regions()
Out[8]:
Index(['AT', 'BE', 'BG', 'CY', 'CZ', 'DE', 'DK', 'EE', 'ES', 'FI', 'FR', 'GR',
'HU', 'IE', 'IT', 'LT', 'LU', 'LV', 'MT', 'NL', 'PL', 'PT', 'RO', 'SE',
'SI', 'SK', 'GB', 'US', 'JP', 'CN', 'CA', 'KR', 'BR', 'IN', 'MX', 'RU',
'AU', 'CH', 'TR', 'TW', 'NO', 'ID', 'ZA', 'WA', 'WL', 'WE', 'WF', 'WM'],
dtype='object', name='region')
In [9]:
list(exio2.get_extensions())
Out[9]:
['factor_inputs', 'emissions', 'materials', 'resources', 'impact']
Calculating the system and extension results¶
The following command checks for missing parts in the system and calculates them. In case of the parsed EXIOBASE this includes A, L, multipliers M, footprint accounts, ..
In [10]:
exio2.calc_all()
Out[10]:
<pymrio.core.mriosystem.IOSystem at 0x7f16909c1f98>
Exploring the results¶
In [11]:
import matplotlib.pyplot as plt
plt.figure(figsize=(15,15))
plt.imshow(exio2.A, vmax=1E-3)
plt.xlabel('Countries - sectors')
plt.ylabel('Countries - sectors')
plt.show()
The available impact data can be checked with:
In [12]:
list(exio2.impact.get_rows())
Out[12]:
['Value Added',
'Employment',
'Employment hour',
'abiotic depletion (elements, ultimate ultimate reserves)',
'abiotic depletion (fossil fuels)',
'abiotic depletion (elements, reserve base)',
'abiotic depletion (elements, economic reserve)',
'Landuse increase of land competition',
'global warming (GWP100)',
'global warming net (GWP100 min)',
'global warming net (GWP100 max)',
'global warming (GWP20)',
'global warming (GWP500)',
'ozone layer depletion (ODP steady state)',
'ozone layer depletion (ODP5)',
'ozone layer depletion (ODP10)',
'ozone layer depletion (ODP15)',
'ozone layer depletion (ODP20)',
'ozone layer depletion (ODP25)',
'ozone layer depletion (ODP30)',
'ozone layer depletion (ODP40)',
'human toxicity (HTP inf)',
'Freshwater aquatic ecotoxicity (FAETP inf)',
'Marine aquatic ecotoxicity (MAETP inf)',
'Freshwater sedimental ecotoxicity (FSETP inf)',
'Marine sedimental ecotoxicity (MSETP inf)',
'Terrestrial ecotoxicity (TETP inf)',
'human toxicity (HTP20)',
'Freshwater aquatic ecotoxicity (FAETP20)',
'Marine aquatic ecotoxicity (MAETP20)',
'Freshwater sedimental ecotoxicity (FSETP20)',
'Marine sedimental ecotoxicity (MSETP20)',
'Terrestrial ecotoxicity (TETP20)',
'human toxicity (HTP100)',
'Freshwater aquatic ecotoxicity (FAETP100)',
'Marine aquatic ecotoxicity (MAETP100)',
'Freshwater sedimental ecotoxicity (FSETP100)',
'Marine sedimental ecotoxicity (MSETP100)',
'Terrestrial ecotoxicity (TETP100)',
'human toxicity (HTP500)',
'Freshwater aquatic ecotoxicity (FAETP500)',
'Marine aquatic ecotoxicity (MAETP500)',
'Freshwater sedimental ecotoxicity (FSETP500)',
'Marine sedimental ecotoxicity (MSETP500)',
'Terrestrial ecotoxicity (TETP500)',
'Human toxicity (USEtox) 2008',
'Fresh water Ecotoxicity (USEtox) 2008',
'Human toxicity (USEtox) 2010',
'Fresh water Ecotoxicity (USEtox) 2010',
'photochemical oxidation (high NOx)',
'photochemical oxidation (low NOx)',
'photochemical oxidation (MIR; very high NOx)',
'photochemical oxidation (MOIR; high NOx)',
'photochemical oxidation (EBIR; low NOx)',
'acidification (incl. fate, average Europe total, A&B)',
'acidification (fate not incl.)',
'eutrophication (fate not incl.)',
'eutrophication (incl. fate, average Europe total, A&B)',
'radiation',
'odour',
'EPS',
'Carcinogenic effects on humans (H.A)',
'Respiratory effects on humans caused by organic substances (H.A)',
'Respiratory effects on humans caused by inorganic substances (H.A)',
'Damages to human health caused by climate change (H.A)',
'Human health effects caused by ionising radiation (H.A)',
'Human health effects caused by ozone layer depletion (H.A)',
'Damage to Ecosystem Quality caused by ecotoxic emissions (H.A)',
'Damage to Ecosystem Quality caused by the combined effect of acidification and eutrophication (H.A)',
'Damage to Ecosystem Quality caused by land occupation (H.A)',
'Damage to Ecosystem Quality caused by land conversion (H.A)',
'Damage to Resources caused by extraction of minerals (H.A)',
'Damage to Resources caused by extraction of fossil fuels (H.A)',
'Carcinogenic effects on humans (E.E)',
'Respiratory effects on humans caused by organic substances (E.E)',
'Respiratory effects on humans caused by inorganic substances (E.E)',
'Damages to human health caused by climate change (E.E)',
'Human health effects caused by ionising radiation (E.E)',
'Human health effects caused by ozone layer depletion (E.E)',
'Damage to Ecosystem Quality caused by ecotoxic emissions (E.E))',
'Damage to Ecosystem Quality caused by the combined effect of acidification and eutrophication (E.E)',
'Damage to Ecosystem Quality caused by land occupation (E.E)',
'Damage to Ecosystem Quality caused by land conversion (E.E)',
'Damage to Resources caused by extraction of minerals (E.E)',
'Damage to Resources caused by extraction of fossil fuels (E.E)',
'Carcinogenic effects on humans (I.I)',
'Respiratory effects on humans caused by organic substances (I.I)',
'Respiratory effects on humans caused by inorganic substances (I.I)',
'Damages to human health caused by climate change (I.I)',
'Human health effects caused by ionising radiation (I.I)',
'Human health effects caused by ozone layer depletion (I.I)',
'Damage to Ecosystem Quality caused by ecotoxic emissions (I.I)',
'Damage to Ecosystem Quality caused by the combined effect of acidification and eutrophication (I.I)',
'Damage to Ecosystem Quality caused by land occupation (I.I)',
'Damage to Ecosystem Quality caused by land conversion (I.I)',
'Damage to Resources caused by extraction of minerals (I.I)',
'Damage to Resources caused by extraction of fossil fuels (I.I)',
'photochemical oxidation (high NOx)(incl. NOx average, NMVOC average)',
'photochemical oxidation (high NOx)(incl. NMVOC average)',
'human toxicity HTP inf. (incl. PAH average, Xylene average, NMVOC average)',
'Freshwater aquatic ecotoxicity FAETP inf. (incl. PAH average, Xylene average, NMVOC average)',
'Marine aquatic ecotoxicity MAETP inf. (incl. PAH average, Xylene average, NMVOC average)',
'Terrestrial ecotoxicity TETP inf. (incl. PAH average, Xylene average, NMVOC average)',
'global warming GWP100 (incl. NMVOC average)',
'ozone layer depletion ODP steady state (incl. NMVOC average)',
'Total Emission relevant energy use',
'Total Energy inputs from nature',
'Total Energy supply',
'Total Energy Use',
'Total Heat rejected to fresh water',
'Domestic Extraction',
'Unused Domestic Extraction',
'Water Consumption Green - Agriculture',
'Water Consumption Blue - Agriculture',
'Water Consumption Blue - Livestock',
'Water Consumption Blue - Manufacturing',
'Water Consumption Blue - Electricity',
'Water Consumption Blue - Domestic',
'Water Consumption Blue - Total',
'Water Withdrawal Blue - Manufacturing',
'Water Withdrawal Blue - Electricity',
'Water Withdrawal Blue - Domestic',
'Water Withdrawal Blue - Total',
'Land use']
And to get for example the footprint of a specific impact do:
In [13]:
print(exio2.impact.unit.loc['global warming (GWP100)'])
exio2.impact.D_cba_reg.loc['global warming (GWP100)']
unit kg CO2 eq.
Name: global warming (GWP100), dtype: object
Out[13]:
region
AT 1.450787e+11
BE 1.991422e+11
BG 6.266676e+10
CY 1.556996e+10
CZ 1.471491e+11
DE 1.394892e+12
DK 1.079304e+11
EE 2.381673e+10
ES 6.079175e+11
FI 1.153875e+11
FR 8.019998e+11
GR 2.247927e+11
HU 9.096635e+10
IE 9.591233e+10
IT 8.419421e+11
LT 3.366823e+10
LU 1.467799e+10
LV 2.255212e+10
MT 5.014763e+09
NL 2.992112e+11
PL 4.136385e+11
PT 1.120749e+11
RO 1.543358e+11
SE 1.282029e+11
SI 3.239223e+10
SK 6.911104e+10
GB 1.073548e+12
US 7.591895e+12
JP 1.825128e+12
CN 6.986984e+12
CA 7.142173e+11
KR 7.566406e+11
BR 5.595118e+11
IN 1.658771e+12
MX 6.219372e+11
RU 1.635710e+12
AU 5.715893e+11
CH 1.201448e+11
TR 4.939783e+11
TW 2.924074e+11
NO 8.791708e+10
ID 4.552600e+11
ZA 3.547961e+11
WA 1.224565e+12
WL 7.970228e+11
WE 4.931660e+11
WF 6.100073e+11
WM 1.329488e+12
Name: global warming (GWP100), dtype: float64
Visualizing the data¶
In [14]:
with plt.style.context('ggplot'):
exio2.impact.plot_account(['global warming (GWP100)'], figsize=(15,10))
plt.show()
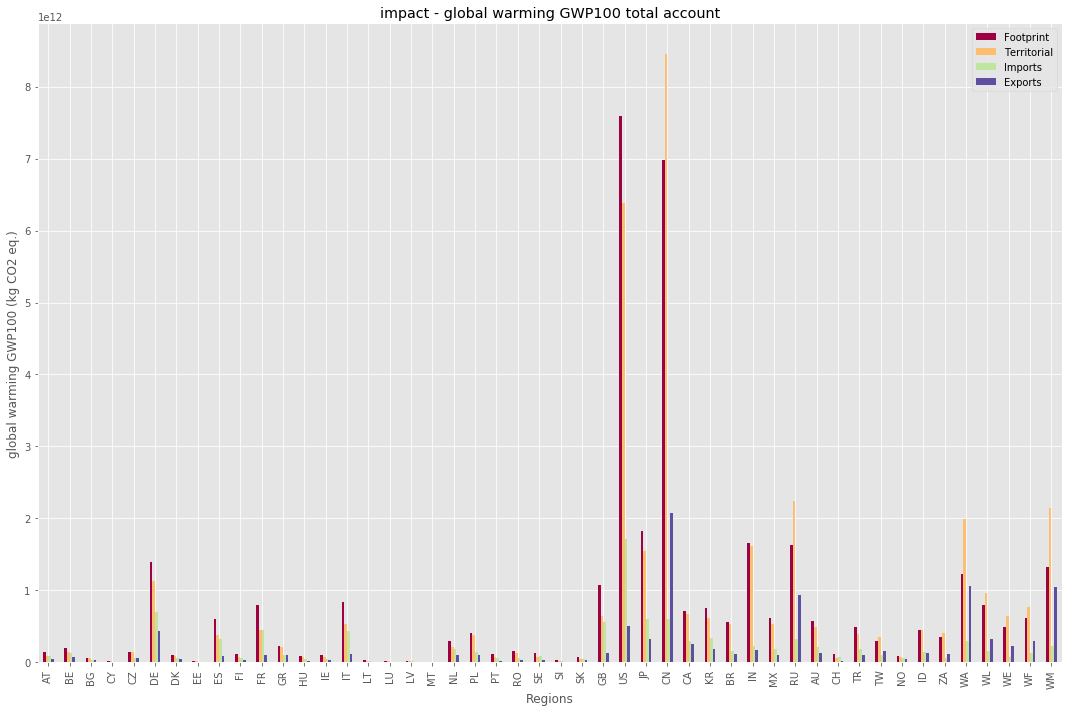
See the other notebooks for further information on aggregation and file io.